Common Issues
I am getting the error message: “Could not load a valid TreeBuilder (ILP solvers), tried ‘[GUROBI, CPLEX, CLP, GLPK]’. Please read the installation instructions.”
SIRIUS uses an ILP solver to calculate fragmentation trees. It ships with the non-commercial CLP solver and should work out-of-the box. If you are nevertheless experiencing this issues, please contact us. It is almost impossible to reproduce and fix such issues without additional information from users. Thanks.
For more information on ILP solvers see the installation instructions.
How to configure SIRIUS to compute large data sets that contain high mass compounds?
When computing molecular formulas with SIRIUS a few high mass compounds usually need most of the computing time, and some of them might not finish computing in reasonable time at all and block your whole analysis.
The most straightforward solution is to exclude high mass compounds from the analysis,
by setting a mass threshold. This will usually allow you to annotate the vast majority of your data. However, many of
the higher mass compounds will work just fine, and it would be a pity to not annotate them. Therefore, the recommended
solution is to set a per-compound timeout (--compound-timeout) so that a few hard cases cannot block your analysis.
Using SIRIUS
How can I export result in SIRIUS 4.4 GUI?
SIRIUS 4.4 saves all results and data immediately to disk. This output is a well organized folder structure (project-space).
If you have not specified any custom location (SaveAs button), SIRIUS stores everything in a temporary location per default.
You can see the path of current project-space location in the header of the SIRIUS window.
How can I access the Probability of ClassyFire classes predicted by CANOPUS
In your project space directory there should be a canopus.tsv file.
This file lists all compound classes with meta information, and their relative index.
The relative index (starting with 0) tells you which line in the canopus .fpt files belongs to which compound class.
Alternatively, you can use the canopus_treemap python library which contains code for parsing the compound classes from the project space.
How should I interpret COSMIC confidence values?
COSMIC confidence scores need to be interpreted with some care. It is important to understand, that they do not represent probabilities, and thus there is no statistical interpretation of a confidence value. If doing large scale analysis, one should focus on the highest confident hits (for example the Top 1/5/10%), (mostly) regardless of their confidence value.
An annotation received a low COSMIC score, even though am I sure it is correct!
In this example, the spectrum of Campherol from the SIRIUS demo data receives the low COSMIC score of 0.3, even though the structure annotation is correct. The reason for this is, that the database we searched in contains multiple extremely similar structures, that have very similar fingerprint representations as well as CSI:FingerID scores. If the true structure was not know, each of these highly-similar compounds could be the correct structure. Hence, the hit receives a low confidence. This is not a limitation of CSI:FingerID and COSMIC but rather of mass spectrometry. The MS/MS of these structures will look very similar.
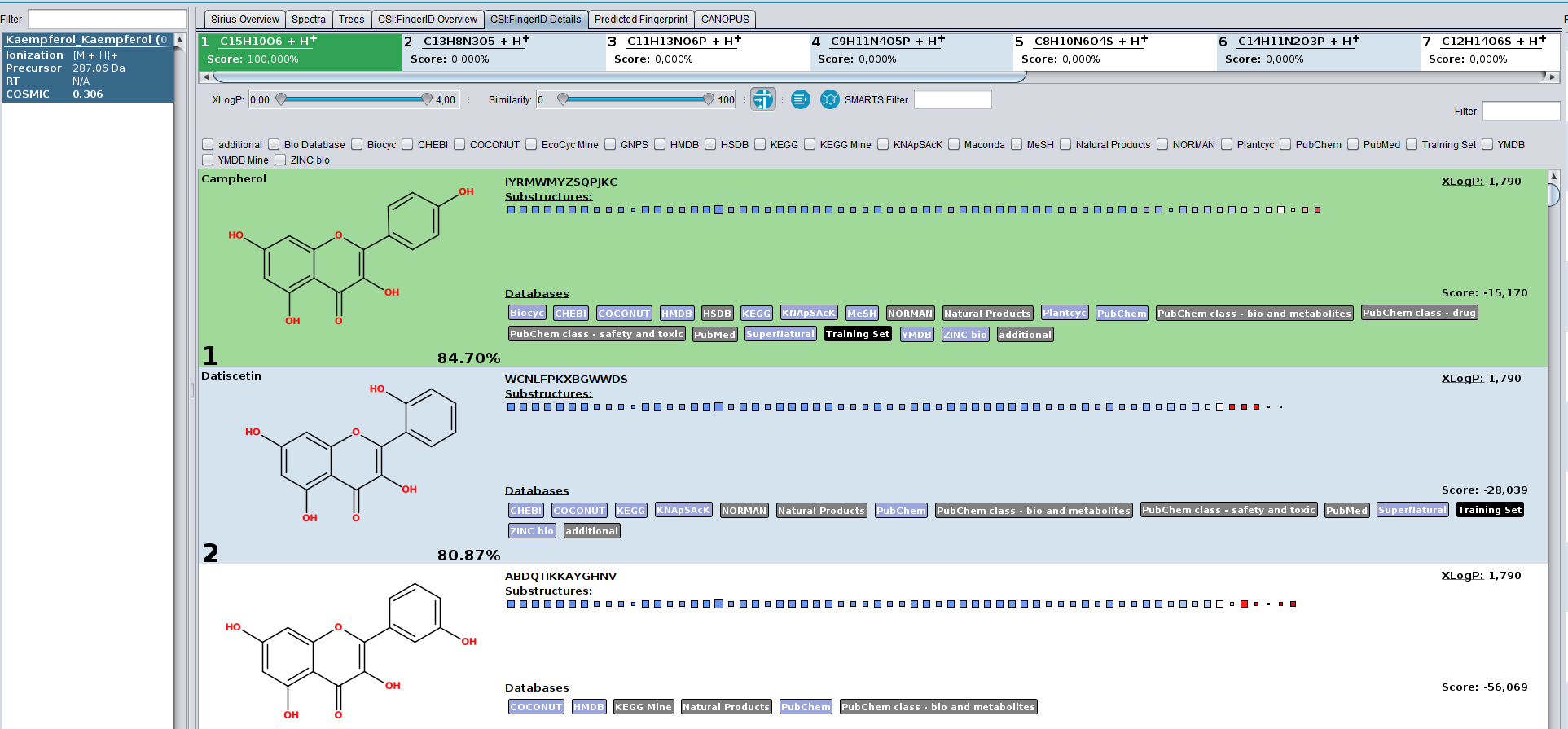
COSMIC Campherol.
Feature Requests
Could SIRIUS support for multiple charged ions?
Not in the short term since there is a plenty of algorithmic work that has to be done. Further, we do not have enough training data of multiple charged ions. If you can provide such data this would help a lot to get such project started.
Could SIRIUS support for Multimeres?
Yes it possible in principle but it needs some adaptions of the fragmentation tree computation that are not implemented yet. It is on our list for future features.